def finite_difference(X, dt):
n_vars, n_samples = X.shape
# Use central differences where possible, forward/backward at boundaries
dXdt = np.zeros((n_samples, n_vars))
# Forward difference at first point
dXdt[0, :] = (X[:, 1] - X[:, 0]) / dt
# Central differences for interior points
for i in range(1, n_samples - 1):
dXdt[i, :] = (X[:, i + 1] - X[:, i - 1]) / (2 * dt)
# Backward difference at last point
dXdt[-1, :] = (X[:, -1] - X[:, -2]) / dt
return dXdt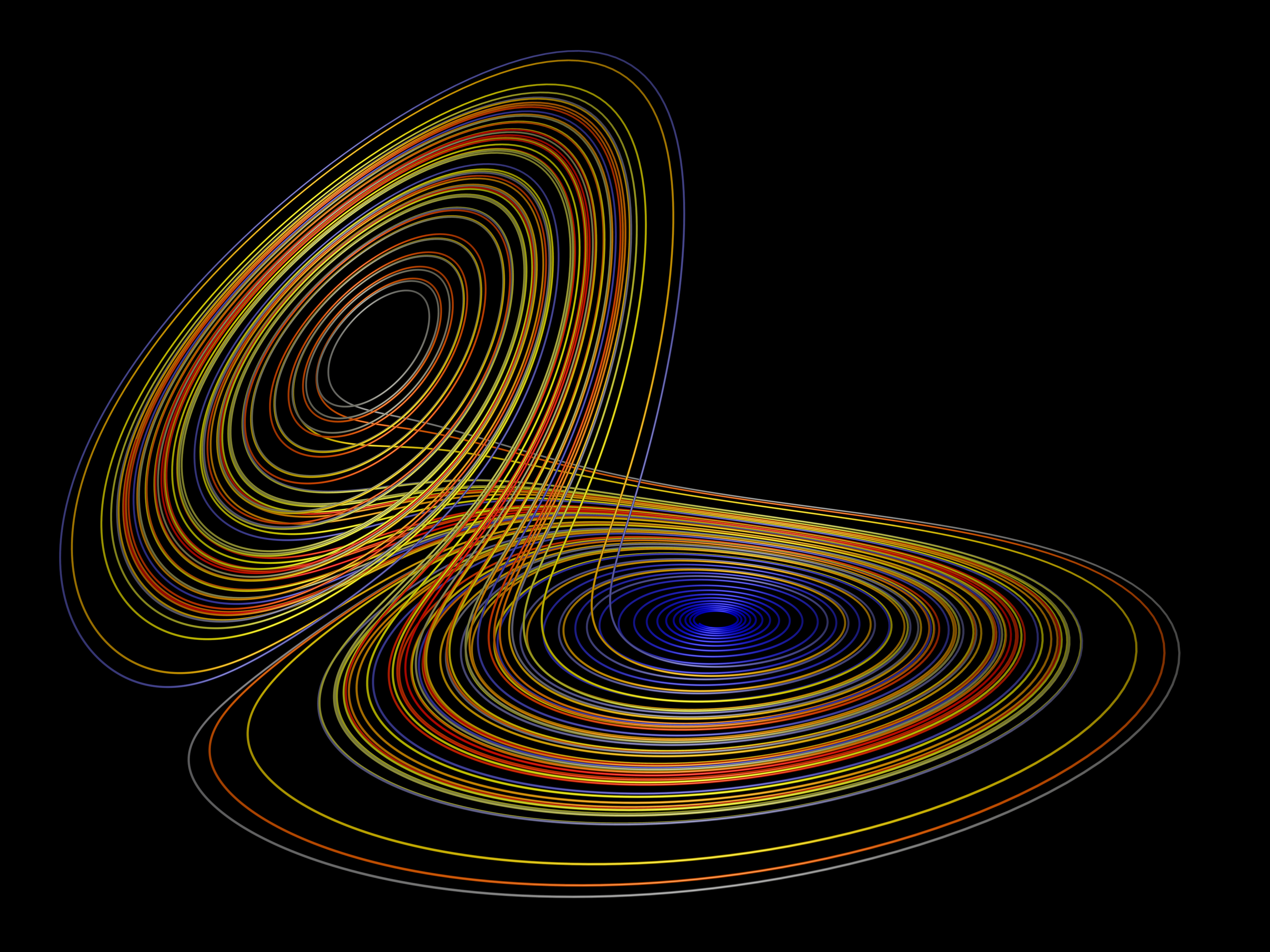
Introduction
In the last post I looked at DMD and EDMD, two methods for analyzing dynamical systems. Both methods look at pairs of snapshots \((x_k, x_{k+1})\) and fit a linear update rule that best predicts the next snapshot. By inspecting that linear map’s eigenvalues and eigenvectors we can learn which patterns dominate and how fast they grow or decay.
Unfortunately, both of these methods rely on a set of features you have chosen in advance. Without the right features you might miss important physics; too many features and the model will become unwieldy. Additionally, while the spectral methods provide some useful insight, they might not have the most easily interpretable output.
What we need is a sparse method, so that we can test a wide number of features and only select the relevant ones for the dynamics, ideally to recover an actual set of interpretable governing equations.
Sparse Identification of Nonlinear Dynamical Systems (SINDy)
SINDy creates a library of candidate basis functions for the eigenfunctions (like EDMD), then does an L1-regularized1 regression to determine which ones to use. There are two formulations: discrete-time and continuous-time.
Discrete Time
This setup is the same as in DMD and EDMD. We have a discrete-time dynamical system of form:
\[ x_{t+1} = F(x_t) \]
We have our data stream, which we use to create our matrix \(X\):
\[ \mathbf{X} = \begin{bmatrix} \mathbf{x}^T(t_0) \\ \mathbf{x}^T(t_1) \\ \vdots \\ \mathbf{x}^T(t_{m-1}) \end{bmatrix} = \begin{bmatrix} x_1(t_0) & x_2(t_0) & \cdots & x_n(t_0) \\ x_1(t_1) & x_2(t_1) & \cdots & x_n(t_1) \\ \vdots & \vdots & \ddots & \vdots \\ x_1(t_{m-1}) & x_2(t_{m-1}) & \cdots & x_n(t_{m-1}) \end{bmatrix} \]
and its time-shifted counterpart \(Y\):
\[ \mathbf{Y} = \begin{bmatrix} \mathbf{x}^T(t_1) \\ \mathbf{x}^T(t_2) \\ \vdots \\ \mathbf{x}^T(t_m) \end{bmatrix} = \begin{bmatrix} x_1(t_1) & x_2(t_1) & \cdots & x_n(t_1) \\ x_1(t_2) & x_2(t_2) & \cdots & x_n(t_2) \\ \vdots & \vdots & \ddots & \vdots \\ x_1(t_m) & x_2(t_m) & \cdots & x_n(t_m) \end{bmatrix} \]
Now, we need some “library matrix” of relevant basis functions applied to the data (similar to EDMD). This may look something like:
\[ \mathbf{\Theta}(\mathbf{X}) := [\theta_1(X), \theta_2(X),...,\theta_{\ell}(X)] \]
This matrix can be quite large. For example, if the basis functions are drawn from combinations of the monomials \(x_1\), \(x_2\), and \(x_3\) (up to quadratic), we would have something like:
\[ \Theta(X) = \begin{bmatrix} | & | & | & | & | & | & & | \\ 1 & x_1 & x_2 & x_3 & x_1^2 & x_1x_2 & \cdots & x_3^2 \\ | & | & | & | & | & | & & | \end{bmatrix} \]
We are looking for a sparse set of coefficients \(\mathbf{\Xi}\) such that
\[ \mathbf{\Xi} = \begin{bmatrix} | & | & & | \\ \xi_1 & \xi_2 & \cdots & \xi_n \\ | & | & & | \end{bmatrix} \]
This leads us to the SINDy equation:
\[ \mathbf{Y} = \mathbf{\Theta}(\mathbf{X}) \mathbf{\Xi} \]
Each basis function is weighted by a \(\xi_i\). To find the \(\xi_i\), we run a sparse \(L1\)-regularized regression:
\[ \Xi_{opt} = \text{arg }\text{min}_{\Xi} ||\mathbf{Y} - \mathbf{\Theta}(\mathbf{X})\mathbf{\Xi} ||_{2} + \mu||\mathbf{\Xi}||_{1} \]
where \(\mu\) is a parameter controlling the strength of the regularization.
To see the mapping at a single time-step, take the \(k\)-th row of \(\Theta(X)\):
\[ \boxed{\;x^{(k+1)\top} = \Theta\!\bigl(x^{(k)}\bigr)\,\Xi\;}, \quad k=0,\dots,m-1, \]
where
\[ \Theta(x^{(k)})=[\theta_1(x^{(k)}),\dots,\theta_L(x^{(k)})]\in\mathbb R^{1\times L} \]
Continuous Time
A more typical formulation is to assume a dynamical system of the form
\[ \dot{x} = \frac{dx}{dt} = f(x(t)) \]
where \(x\) is the state (possibly a vector) and \(t\) is the time. We have moved from discrete-time to continuous time. We now seek to learn the vector field \(f\), which relates the current state to the rate of change in the state, rather than the next state. Luckily, as we describe in the previous post the Koopman eigenfunctions satisfy:
\[ \frac{d}{dt}\phi(x) = K\phi(x) = \lambda \phi(x) \]
By the chain rule, we also have
\[ \frac{d}{dt}\phi(x) = \nabla \phi(x) \cdot \dot{x} = \nabla \phi(x) \cdot f(x) \]
Combining the two, we have
\[ \nabla \phi(x) \cdot f(x) = \lambda \phi(x) \]
So we can approximate the eigenfunctions via regression2. Approximate \(f(x)\) by a sparse weighted sum of dictionary elements:
\[ f(x)\;=\;\sum_{j=1}^{L}\xi_{j}\,\theta_{j}(x)\;=\;\mathbf{\Theta}(x)\,\mathbf{\Xi} \]
This gives:
\[ \nabla\phi(x)\,\cdot\,\mathbf{\Theta}(x)\,\mathbf{\Xi} \;=\;\lambda\,\phi(x) \]
Evaluating at the sample states:
\[ \nabla\phi\!\bigl(\mathbf{x}^{(i)}\bigr)\, \cdot\,\mathbf{\Theta}\!\bigl(\mathbf{x}^{(i)}\bigr)\, \mathbf{\Xi} \;=\; \lambda\, \phi\!\bigl(\mathbf{x}^{(i)}\bigr), \quad i=1,\dots,m \]
Now we have
\[ \mathbf{\dot X} \;=\; \mathbf{\Theta}(\mathbf{X})\,\mathbf{\Xi}, % (5) \quad\text{where} \; \mathbf{\dot X} = \begin{bmatrix} \dot{\mathbf x}^{(1)\!\top}\\ \dot{\mathbf x}^{(2)\!\top}\\ \vdots\\ \dot{\mathbf x}^{(m)\!\top} \end{bmatrix},\; \mathbf{\Theta}(\mathbf{X}) = \begin{bmatrix} \mathbf{\Theta}(x^{(1)})\\ \mathbf{\Theta}(x^{(2)})\\ \vdots\\ \mathbf{\Theta}(x^{(m)}) \end{bmatrix} \]
and so we can solve the continuous-time problem with the following regression
\[ \mathbf{\Xi}_{opt} \;=\; \operatorname*{arg\,min}_{\mathbf{\Xi}} \Bigl\| \mathbf{\dot X} - \mathbf{\Theta}(\mathbf{X})\,\mathbf{\Xi} \Bigr\|_{2} \;+\; \mu\,\|\mathbf{\Xi}\|_{1} \]
Conveniently, exactly the same as the previous regression equation, except here we are approximating:
\[ \frac{dx}{dt} = f(x(t)) = \mathbf{\Theta}(x)\mathbf{\Xi} \]
Implementation
To implement SINDy, we need a library matrix function and a method of handling the regression. We may also want a finite differences method, to calculate derivatives.
Finite Differences
We will use finite differences3 to approximate the time derivative by dividing small state increments by the timestep \(\Delta t\).
At an interior point \(t_i\) we use the second-order central stencil
\[
\dot x(t_i)\;\approx\;\frac{x(t_{i+1}) - x(t_{i-1})}{2\,\Delta t},
\]
which cancels the first-order truncation error, yielding \(\mathcal{O}(\Delta t^{2})\) accuracy.
At the boundaries we fall back to the first-order forward and backward stencils: \[ \dot x(t_0)\;\approx\;\frac{x(t_1)-x(t_0)}{\Delta t}, \qquad \dot x(t_{m-1})\;\approx\;\frac{x(t_{m-1})-x(t_{m-2})}{\Delta t}. \]
The helper above implements exactly this scheme: it accepts a state matrix of shape \((n_{\text{vars}}, n_{\text{samples}})\), returns the derivative matrix \((n_{\text{samples}}, n_{\text{vars}})\), and also transposes \(X\) so rows correspond to time snapshots for the subsequent regression.
Library Matrix
Next we build the candidate feature matrix \(\Theta\) by stacking a constant term, all monomials up to poly_order, and optionally sine/cosine transforms of each state variable. Returns that matrix along with a parallel descriptions list so the sparse coefficients can later be mapped back to human-readable terms.
def sindy_library(X, poly_order=3, include_sine=False, include_cosine=False):
n_vars, n_samples = X.shape
# Start with constant term
library_functions = [np.ones(n_samples)]
descriptions = ['1']
# Add polynomial terms
for order in range(1, poly_order + 1):
for combo in combinations_with_replacement(range(n_vars), order):
if order == 1:
var_idx = combo[0]
library_functions.append(X[var_idx, :])
descriptions.append(f'x_{var_idx}')
else:
term = np.ones(n_samples)
term_desc = []
for var_idx in combo:
term *= X[var_idx, :]
term_desc.append(f'x_{var_idx}')
library_functions.append(term)
descriptions.append('*'.join(term_desc))
# Add trigonometric terms if requested
if include_sine:
for i in range(n_vars):
library_functions.append(np.sin(X[i, :]))
descriptions.append(f'sin(x_{i})')
if include_cosine:
for i in range(n_vars):
library_functions.append(np.cos(X[i, :]))
descriptions.append(f'cos(x_{i})')
Theta = np.column_stack(library_functions)
return Theta, descriptionsSequential Threshold Least Squares
We will use Sequential Threshold Least Squares to obtain the full coefficient matrix.
At each iteration \(k\):
Set every entry with \(|\xi_{ij}^{(k)}| < \lambda_{reg}\) to zero, forcing small terms out of the model4.
Refit for each state component \(j\), solve a new least–squares problem using only the surviving (non-zero) columns of \(\Theta\) to get updated weights.
Repeat until convergence or for at most
max_itercycles; the final \(\Xi\) contains only the terms that remain large after repeated shrink-and-refit, giving a sparse governing equation.
def sequential_threshold_least_squares(Theta, dXdt, lambda_reg=0.1, max_iter=10):
n_states = dXdt.shape[1]
# Initialize coefficient matrix
Xi = np.linalg.lstsq(Theta, dXdt, rcond=None)[0]
# Iterative thresholding
for iteration in range(max_iter):
# Find small coefficients to remove
small_inds = np.abs(Xi) < lambda_reg
# Set small coefficients to zero
Xi[small_inds] = 0
# Identify active (non-zero) coefficients for each state
for i in range(n_states):
big_inds = ~small_inds[:, i]
if np.any(big_inds):
# Recompute non-zero coefficients using least squares
Xi[big_inds, i] = np.linalg.lstsq(
Theta[:, big_inds], dXdt[:, i], rcond=None
)[0]
else:
Xi[:, i] = 0
return Xiprint_equations
I’ll add one bonus helper function, which will be useful later
def print_equations(Xi, descriptions, feature_names=None):
n_functions, n_features = Xi.shape
if feature_names is None:
feature_names = [f'x_{i}' for i in range(n_features)]
for i in range(n_features):
# Build equation string
terms = []
for j in range(n_functions):
coef = Xi[j, i]
if abs(coef) > 1e-10: # Only include non-zero terms
if abs(coef - 1.0) < 1e-10:
terms.append(f"{descriptions[j]}")
elif abs(coef + 1.0) < 1e-10:
terms.append(f"-{descriptions[j]}")
else:
terms.append(f"{coef:.6f}*{descriptions[j]}")
if terms:
equation = " + ".join(terms).replace(" + -", " - ")
print(f"d{feature_names[i]}/dt = {equation}")
else:
print(f"d{feature_names[i]}/dt = 0")
print()Integration
Putting the piece together:
def sindy(X, dXdt, poly_order=3, lambda_reg=0.1, include_sine=False,
include_cosine=False, max_iter=10):
# Build library of candidate functions
Theta, descriptions = sindy_library(
X,
poly_order=poly_order,
include_sine=include_sine,
include_cosine=include_cosine
)
# Sparse regression using sequential thresholded least squares
Xi = sequential_threshold_least_squares(Theta, dXdt, lambda_reg, max_iter)
return Xi, descriptionsFor a production version you may want to factor the library production out of the SINDy method.
Experiments
Let’s generate data from few known systems and see if SINDy can recover their governing equations.
Simple Harmonic Oscillator
Definition
The simple harmonic oscillator is described by the following second-order ODE:
\[ x'' + \omega^2 x = 0 \]
If we let: \[ \begin{align} x_0 &:= x \\ x_1 &:= \frac{dx}{dt} \end{align} \]
We can then derive
\[ \begin{align} \frac{dx_0}{dt} &= \frac{dx}{dt} = x_1 \\ \frac{dx_1}{dt} &= - \omega^2 x_0 \end{align} \]
So it has an alternative formulation as a set of coupled first-order ODEs.
Approximation
The simple harmonic oscillator has a well-known analytic solution: \[ x = \cos(\omega t) \]
Let
def generate_simple_harmonic_oscillator_data(omega=2.0, t_final=10, dt=0.01):
t = np.arange(0, t_final, dt)
# Analytical solution
x = np.cos(omega * t)
xdot = -omega * np.sin(omega * t)
X_train = np.vstack([x, xdot])
return X_trainThis will give us data for the harmonic oscillator5.
Now we run SINDy on this data:
X = generate_simple_harmonic_oscillator_data()
dXdt = finite_difference(X, dt)
Xi, desc = sindy(X, dXdt, poly_order=1, lambda_reg=0.1)
print_equations(Xi, desc, feature_names=['x_0', 'x_1'])We discover the following equations:
dx_0/dt = 0.999925*x_1
dx_1/dt = -3.999764*x_0Which is close to the true generating equations:
\[ \begin{align} \frac{dx_0}{dt} &= x_1 \\ \frac{dx_1}{dt} &= -4 x_0 \end{align} \]
Lorenz System
Definition
The Lorenz-63 model is a three-dimensional ODE derived from a truncated Fourier expansion of the Boussinesq equations for thermal convection. In its nondimensional form the state \(\mathbf{x}=(x,y,z)^{\!\top}\) evolves as
\[ \begin{aligned} \dot x &= \sigma \,(y - x), \\ \dot y &= x \,(\rho - z) - y, \\ \dot z &= x\,y - \beta\,z, \end{aligned} \]
Let us recover this equation from data.
Approximation
def generate_lorenz_data(initial_conditions, sigma=10, rho=28, beta=8/3, t_final=10, dt=0.01):
def lorenz_rhs(state, sigma=sigma, rho=rho, beta=beta):
x, y, z = state
return np.array([
sigma * (y - x),
x * (rho - z) - y,
x * y - beta * z
])
# Generate training data
t_train = np.arange(0, t_final, dt)
n_steps = len(t_train)
X_train = np.zeros((3, n_steps))
X_train[:, 0] = initial_conditions
for i in range(1, n_steps):
k1 = lorenz_rhs(X_train[:, i-1])
k2 = lorenz_rhs(X_train[:, i-1] + dt/2 * k1)
k3 = lorenz_rhs(X_train[:, i-1] + dt/2 * k2)
k4 = lorenz_rhs(X_train[:, i-1] + dt * k3)
X_train[:, i] = X_train[:, i-1] + dt/6 * (k1 + 2*k2 + 2*k3 + k4)
return X_trainFollowing the same steps:
initial_conditions = [-8, 8, 27]
X = generate_lorenz_data(initial_conditions)
dXdt = finite_difference(X, dt)
Xi, desc = sindy(X, dXdt, poly_order=2, lambda_reg=0.05)
print_equations(Xi, desc, feature_names=['x', 'y', 'z'])We discover the following equations:
dx/dt = -9.971816*x_0 + 9.972789*x_1
dy/dt = 27.823397*x_0 - 0.970096*x_1 - 0.994849*x_0*x_2
dz/dt = -2.658203*x_2 + 0.996862*x_0*x_1Which is once again pretty close to the actual generating equations (rewritten): \[ \begin{align} \frac{dx_0}{dt} &= -10x_0 + 10x_1 \\ \frac{dx_1}{dt} &= 28x_0 - x_1 - x_0x_2 \\ \frac{dx_2}{dt} &= -\frac{8}{3}x_2 + x_0x_1 \end{align} \]
Conclusion
We used SINDy to recover governing equations for two toy problems.
Further Reading
Data-Driven Science and Engineering: Machine Learning, Dynamical Systems, and Control.
Discovering governing equations from data by sparse identification of nonlinear dynamical systems.
Data-driven discovery of coordinates and governing equations.
PySINDy – Sparse Identification of Nonlinear Dynamics in Python — open-source implementation.
Footnotes
Also known as “LASSO” in the ML literature. I’ve seen some sources on SINDy do an L\(0\) regularization here, instead; there are many variants of SINDy. You’ll want to follow the typical machine learning best practices (hold-out data, etc.) to fit the best model.↩︎
Assumes the dynamics are both continuous and differentiable. Some systems are more easily represented in the continuous time format than the discrete-time. In the Brunton & Kutz book they also give an alternative method using Laurent series, and some extensions.↩︎
In production, the error from finite differences is too large and may cause SINDy to blow up. Alternatives like smoothed finite differences are available.↩︎
Despite the similar notation \(\lambda_{reg}\) is unrelated to the Koopman eigenvalues.↩︎
This is toy data. In real life, you should probably whiten your data, nondimensionalize, etc.↩︎